Evolutionary flexibility may help plants adapt to climate change
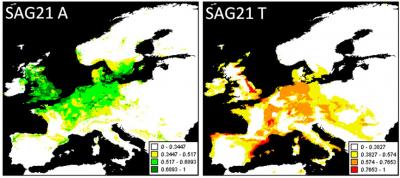 Southern variants of the water stress tolerance gene SAG21 (right panel) were associated with low fitness when grown outside of their home range in the northern Finnish site.In the face of climate change, animals have an advantage over plants: They can move. But a new study shows that plants may have some tricks of their own.
Southern variants of the water stress tolerance gene SAG21 (right panel) were associated with low fitness when grown outside of their home range in the northern Finnish site.In the face of climate change, animals have an advantage over plants: They can move. But a new study shows that plants may have some tricks of their own.
In a paper published in Science, the research team identifies the genetic signature in the common European plant Arabidopsis thaliana that governs the plant’s fitness — its ability to survive and reproduce — in different climates. The researchers further find that climate in large measure influences the suite of genes passed on to Arabidopsis to optimize its survival and reproduction. The set of genes determining fitness varies, the team reports, depending on the climate conditions in the plant’s region — cold, warm, dry, wet, or otherwise.
“This is the first study to show evolutionary adaptation for Arabidopsis thaliana on a broad geographical scale and to link it to molecular underpinnings,” said Johanna Schmitt, director of the Environmental Change Initiative at Brown University and an author on the paper. “Climate is the selective agent.”
The researchers believe that by identifying the genetic signatures that mark Arabidopsis’ response to changing climate, scientists may understand how climate may cause the re-engineering of the genetic profiles of other plants. “There is still evolutionary flexibility to help plants take one direction or another,” said Alexandre Fournier-Level, a postdoctoral researcher at Brown and the paper’s first author. “It gives us good hope to see, yes, it’s adapting.”
The researchers planted Arabidopsis, a small flowering plant popular with plant biologists because its genome is relatively small, at four locations across its native range in Europe — Valencia, Spain; Halle, Germany; Norwich, United Kingdom; and Oulu, Finland. At each field site, genetic strains were planted, originating from across the species’ native climate range — from cold (Finland) to warm (Spain), with oceanic (United Kingdom) and continental (Germany) variables mixed in. That way, the researchers could compare local strains with representatives from the other regions and search for signs of “home court advantage,” Schmitt said.
“This was a truly massive undertaking, tracking more than 75,000 plants in the field, from near the arctic circle to the Mediterranean coast,” said Amity Wilczek, a former postdoctoral researcher in Schmitt’s lab now on the faculty at Deep Springs College. "Arabidopsis is an annual plant, so we could measure total lifetime success of an individual within a single year. We gathered plants from a variety of native climates and grew some of each in our four widely distributed European garden sites. We shipped our harvested plants back to Brown and began the laborious task of counting fruits on these plants. In the end, we were able to assemble a very large and comprehensive dataset that gives us new insight into what it takes for a plant to be succesful in nature under a broad range of climate conditions.”
The team then burrowed into the Arabidopsis genome to find the molecular mechanisms that might give the plant genetic flexibility to roll with climate punches. To identify variations in the genome among the regional representatives, the researchers carried out a genome-wide association study for survival and fruiting comprising more than 213,000 single-nucleotide polymorphisms. These SNPs, Fournier-Level explained, are like signposts pointing to areas in the genome where survival and reproduction may be emphasized and areas that show variations in the regional representatives’ genetic makeup.
From the experiments, the team discovered that the SNPs that determined fitness for Arabidopsis in one region are surprisingly different from those associated with the plant’s fitness in another region. The team also learned from the experiments that SNP variants — “alleles” — associated with high fitness within each field site were locally abundant in that region, demonstrating a kind of home court advantage at the genomic level.
In addition, certain climate variables seemed to control the geographic distribution of fitness-associated SNPs. For example, fitness SNPs in Finland, at the northern range limit, were limited by temperature. In one example presented in the paper, the researchers identify a SNP allele in a water-stress tolerance gene, called SAG21. This allele was common in Arabidopsis’s Spanish populations, but not in the cool climate of Finland where tests showed plants carrying that allele fared poorly.
“Climate explains the distribution of locally favorable alleles,” Fournier-Level explained. “This helps explain how climate shapes distribution.”
“We found that the genetic basis of survival and reproduction is almost entirely different in different regions, which suggests that evolutionary adaptation to one climate may not always result in a tradeoff of poor performance in another climate,” said Schmitt. “Thus, the Arabidopsis genome may contain evolutionary flexibility to respond to climate change.”
Another study of genetic adaptations to climate
In another study published in the same issue of Science, a team led by Joy Bergelson, professor and chair of Ecology and Evolution at the University of Chicago, identified genetic loci associated with adaptations to climate change in A. thaliana.
Genes involved in processes such as photosynthesis and energy metabolism were more common among genes associated with climate adaptation, the researchers discovered. Many of these gene loci also showed evidence of evolving through selective sweeps, where a new mutation appears and spreads through a population — a strategy that may not be effective during rapid changes in climate.
“The contribution of selective sweeps suggests that there will be limits on the rate at which this plant can adapt to climate change,” Bergelson said.
--Reprinted from Brown University




 Climate
Climate
Reader Comments